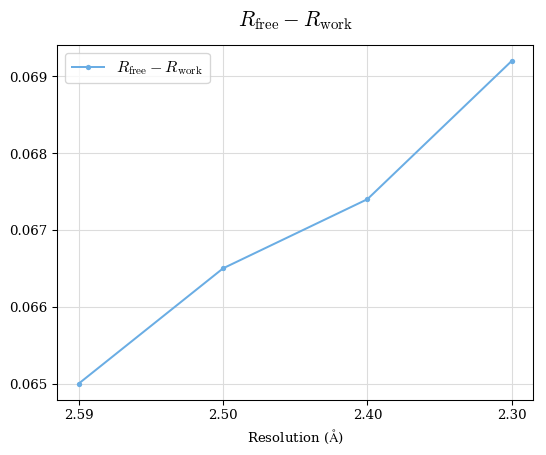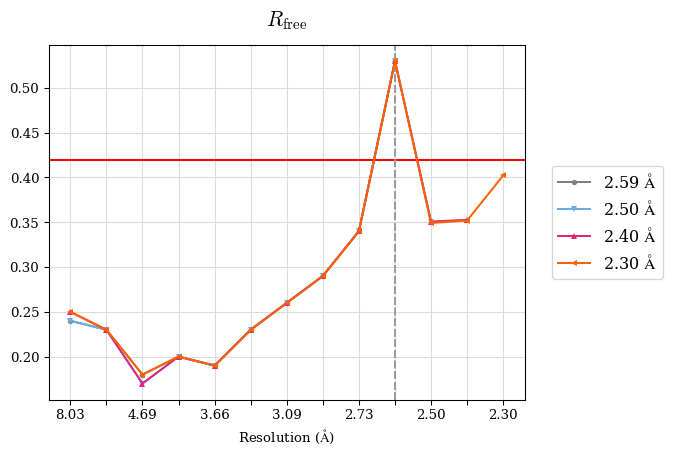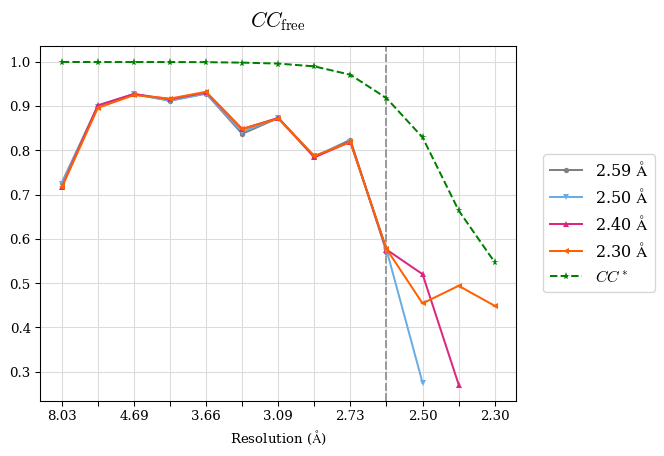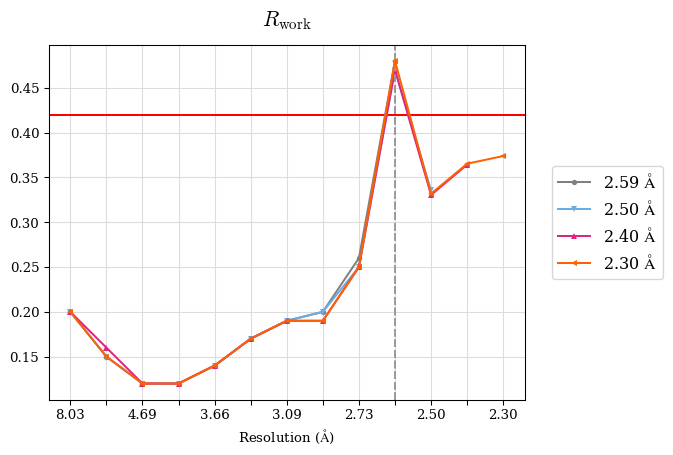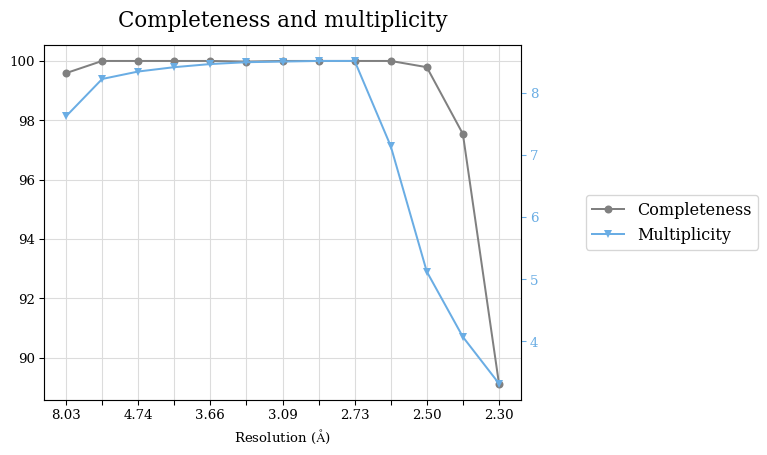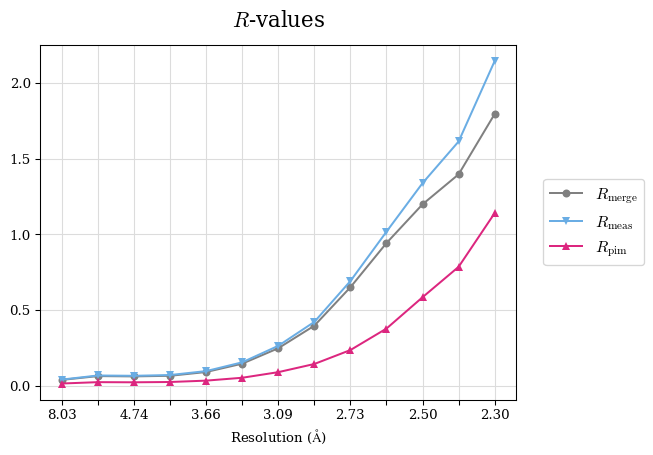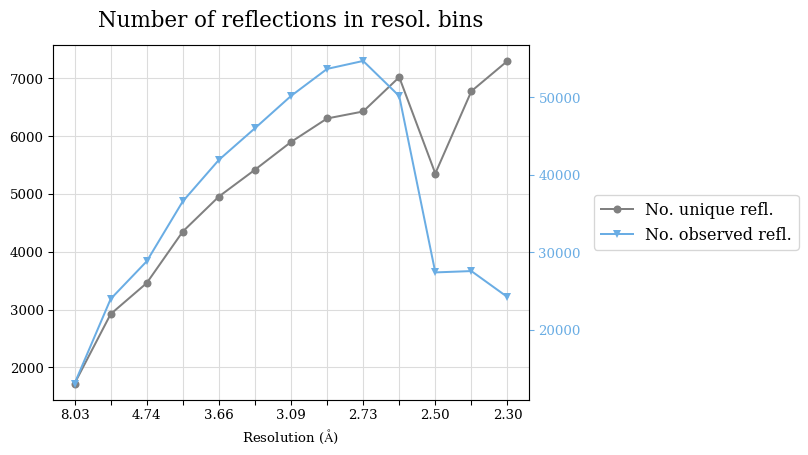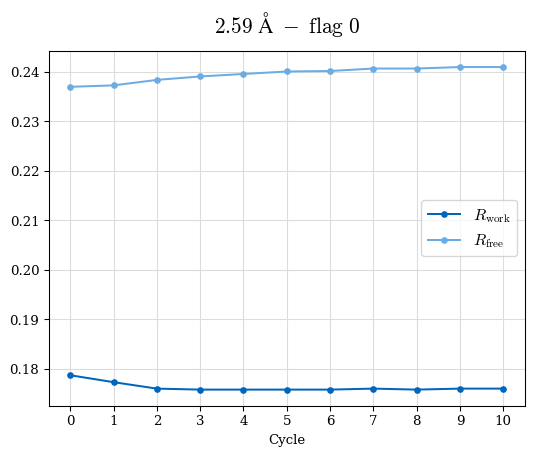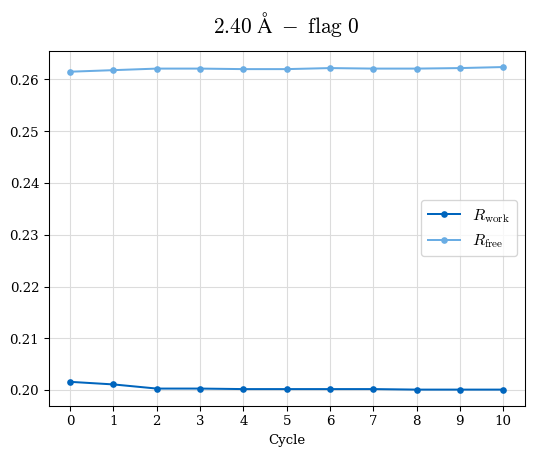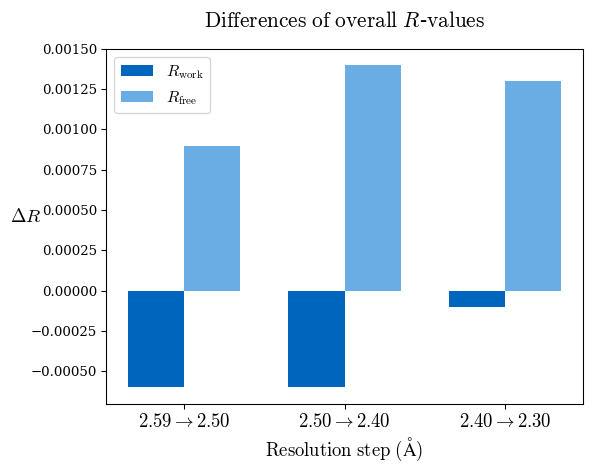
Raw data: BO_R-values.csv
# Shell Rwork(init) Rwork(fin) Rwork(diff) Rfree(init) Rfree(fin) Rfree(diff) 2.59A->2.50A 0.1760 0.1754 -0.0006 0.2410 0.2419 0.0009 2.50A->2.40A 0.1880 0.1874 -0.0006 0.2512 0.2526 0.0014 2.40A->2.30A 0.2001 0.2000 -0.0001 0.2624 0.2637 0.0013
Note: For each incremental step of resolution from X->Y, the R-values were calculated at resolution X.