Automatic PAIRed REFinement protocol
THE WEBSITE HAS BEEN MOVED TO https://pairef.structbio.org. PLEASE CONTINUE THERE.
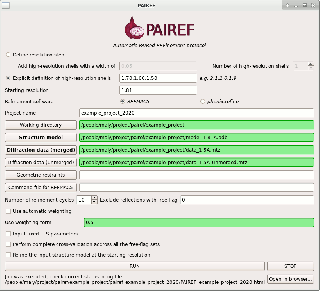
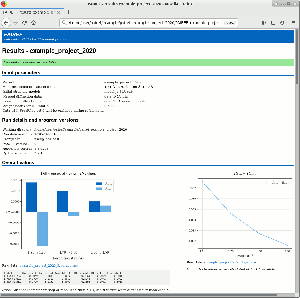
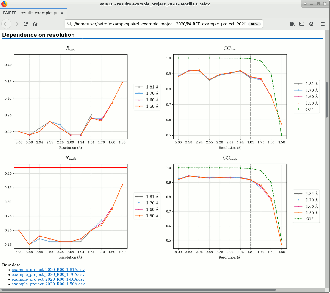
PAIREF is a tool for macromolecular crystallographers that performs the PAIRed REFinement protocol1) automatically to estimate the optimal high-resolution cutoff. It is developed in Python and can be installed as a module into the Computational Crystallography Toolbox. It provides graphical and command-line interface that executes all the needed calculations. Parameters of refinement can be specified in detail to put all the calculations under full control of the user. Obtained results are presented as plots and tables in HTML log file. PAIREF supports REFMAC5 (part of the CCP4 Software Suite) and phenix.refine (part of the PHENIX) for structure model refinement.
Installation and system requirements
PAIREF depends on the installation of the CCP4 Software Suite or PHENIX (both contain Computational Crystallography Toolbox with Python.
PAIREF can be easily installed running the command cctbx.python -m pip install pairef --user --no-deps in terminal (GNU/Linux, macOS) or CCP4Console (Windows). More information are available in documentation. Check also the PAIREF homepage at https://pairef.fjfi.cvut.cz/ and PyPI repository.
Examples
To run paired refinement of a model (previously refined at 1.81 Å) for a series of cutoffs (1.7, 1.6, and 1.5 Å), execute the following command:
cctbx.python -m pairef --XYZIN model_1-81A.pdb --HKLIN data_1-5A.mtz --HKLIN_UNMERGED data_1-5A_unmerged.mtz -i 1.81 -r 1.7,1.6,1.5
For detailed information about other program parameters, read the documentation available at http://pairef.fjfi.cvut.cz/docs.
For the explanation of the paired refinement protocol and a short tutorial, check our video from PAIREF webinar.
Example cases
Example runs presented in our scientific paper Paired refinement under the control of PAIREF (2020) in IUCrJ 7 are available online and can be downloaded from Zenodo:
- simulated data set for lysozyme from Gallus gallus (SIM) – resolution step 0.10 A
- measured data set for thermolysin from Bacillus thermoproteolyticus (TL)
- measured data set for cysteine-bound complex of cysteine dioxygenase from Rattus norvegicus (CDO)
- measured data set for endothiapepsin from Cryphonectria parasitica in complex with fragment B53 (EP) – resolution step 0.05 A
- measured data set for interferon gamma from Paralichthys olivaceus (POLI) – resolution step 0.10 A
- measured data set for bilirubin oxidase from Myrothecium verrucaria (BO)
Publications
M. Maly, K. Diederichs, J. Dohnalek, P. Kolenko: Paired refinement under the control of PAIREF (2020) IUCrJ 7, 681-692 https://doi.org/10.1107/S2052252520005916 PDF
M. Maly, K. Diederichs, J. Dohnalek, P. Kolenko: PAIREF: paired refinement also for Phenix users (2021) Acta Cryst F77, 226-229 https://doi.org/10.1107/S2053230X21006129 PDF
Credits and contact
Please refer: M. Maly, K. Diederichs, J. Dohnalek, P. Kolenko: Paired refinement under the control of PAIREF (2020) IUCrJ 7
PAIREF is developed by Martin Malý in collaboration of Czech Technical University, Czech Academy of Sciences, and University of Konstanz. In case of any questions or problems, please do not hesitate and write us: martin.maly@fjfi.cvut.cz.